
The lit pcba dataset is an important tool for virtual screening. It helps researchers obtain fair and accurate results in their studies. Comprising 149 PubChem bioassays, it eliminates errors and bad data. The dataset includes 9,954 active compounds and 2.7 million inactive ones, making it extremely useful for discovering new medicines. It mimics real experiments to enhance predictions and improve model training. Additionally, it focuses on single protein targets with X-ray structures for practical applications.
Key Takeaways
The LIT-PCBA dataset is important for virtual screening. It gives reliable data similar to real experiments.
It has 149 tests and almost 10,000 active chemicals. This makes it very useful for finding new medicines.
Using LIT-PCBA helps scientists avoid mistakes and make better predictions about how compounds work.
The dataset speeds up the process, letting AI tools work 100 times faster than old methods.
LIT-PCBA is used in drug research for testing virtual screening and training AI models.
What Is the LIT-PCBA Dataset?

Overview and Purpose of the LIT-PCBA Dataset
The LIT-PCBA dataset is a key tool for drug research. It helps scientists test computer models used in virtual screening. Unlike older datasets, it uses high-quality data that feels like real experiments. This makes it great for predicting how proteins and compounds interact. It also improves the accuracy of virtual screening.
The main goal of this dataset is to test how well models work. It uses enrichment factors (EFs) to check if models find active compounds among inactive ones. For instance, the AK-Score-C model had EFs of 6.5 and 4.4 at 0.5% and 1.0% thresholds. These numbers show how useful the dataset is for testing models.
Key Features of the Dataset
The LIT-PCBA dataset has special features that make it helpful for researchers:
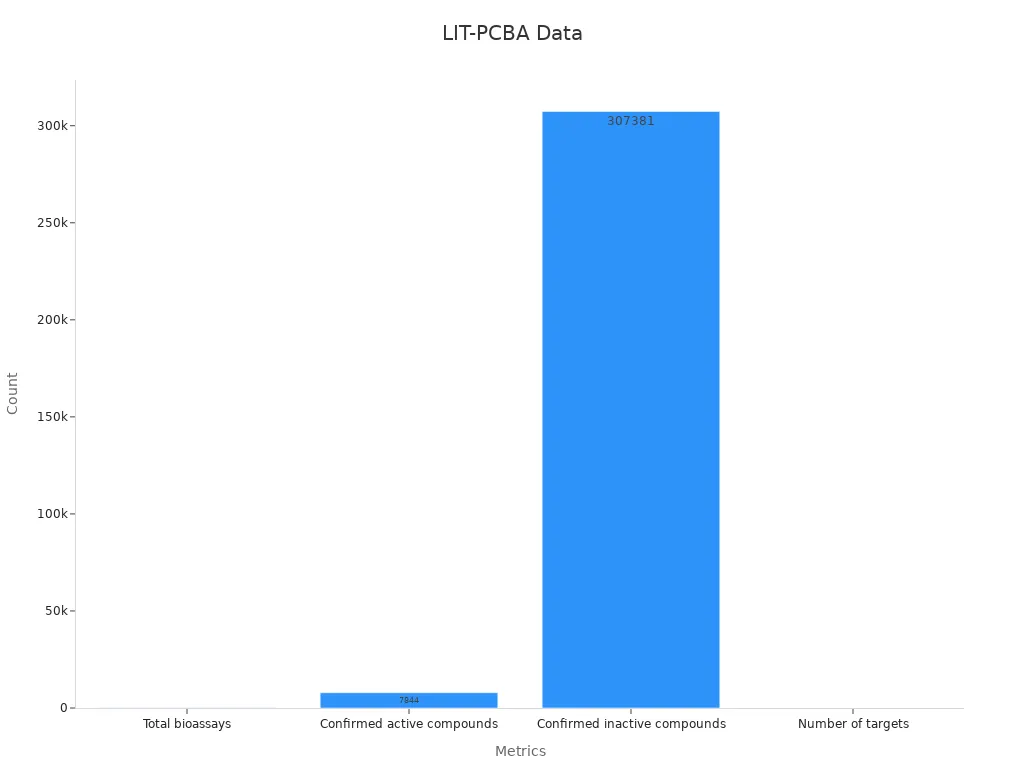
Comprehensive Data: It has 149 bioassays, 7,844 active compounds, and 307,381 inactive ones. This large amount of data helps test models better.
Focus on Single Protein Targets: It studies single protein targets with X-ray structures. This makes it practical for drug discovery.
Unbiased Nature: It uses past data, like the Ames mutagenicity dataset, to stay fair. This helps predict outcomes in pharmaceutical research.
Accelerated Screening: AI tools using this dataset are 100 times faster than older methods. They still keep accurate results.
Here is a table showing key details about the dataset:
Metric | Value |
|---|---|
Total bioassays | 149 |
Confirmed active compounds | 7,844 |
Confirmed inactive compounds | 307,381 |
Number of targets | 15 |
How LIT-PCBA Differs from Other Datasets
The LIT-PCBA dataset fixes problems found in other datasets. Many datasets, like BindingDB and DUD-E, have overlapping data. This overlap makes results less reliable and harder to trust. For example:
Dataset | Issues Identified | Implications for Screening Power |
|---|---|---|
BindingDB | Shares data with other datasets, making results unreliable | Hard to measure overlaps and fairly test screening models |
CASF-2016 | Overlaps with other datasets, making model testing harder | Removing overlaps may lose important protein-ligand data |
DUD-AD | Similar overlap issues, reducing benchmarking accuracy | New models needed to test unseen proteins without bias |
DUD-E | Shares data with others, causing biased evaluations | Fair testing is hard if datasets are changed, affecting comparisons |
LIT-PCBA | Unique but still faces some bias challenges | Shows the need for better models to test binding without bias |
The LIT-PCBA dataset reduces these problems by being more unbiased. Its unique data and validation make it a trusted tool for testing virtual screening models. Researchers prefer it for creating and improving new computer methods.
The Role of LIT-PCBA in Virtual Screening

Why Benchmark Datasets Are Crucial for Virtual Screening
Benchmark datasets are very important for virtual screening. They help test and improve computer models. Using these datasets ensures fair and repeatable results. This makes it easier to compare tools and find the best ones.
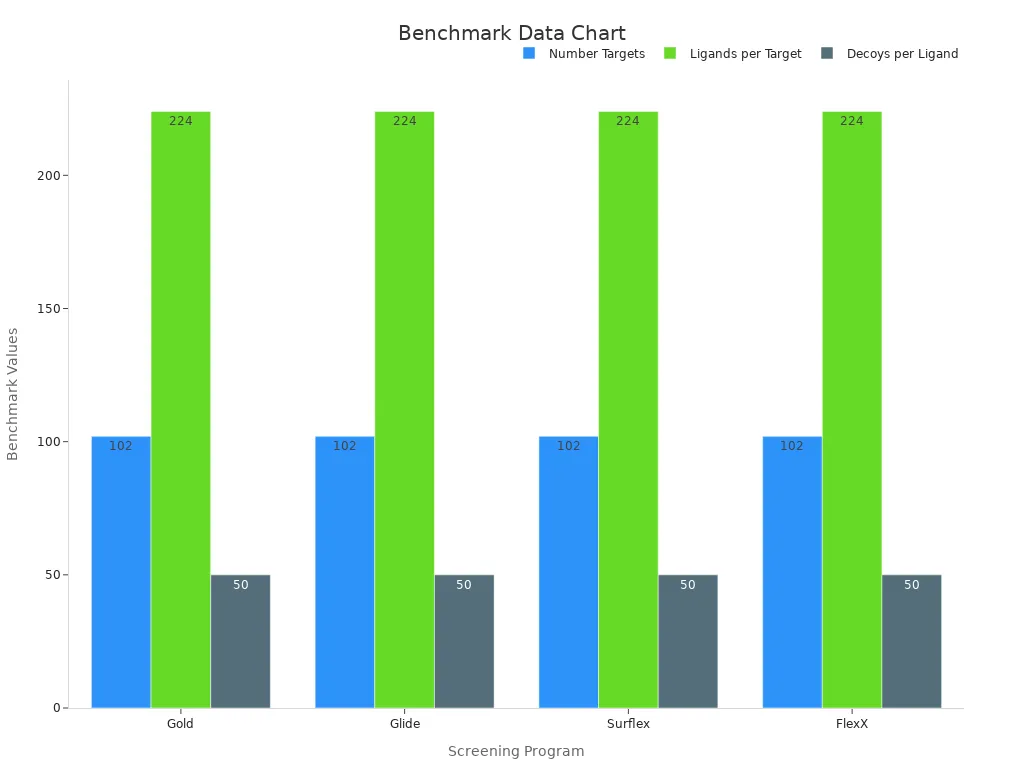
For example, DUD-E is often used in virtual screening programs. Tools like Gold, Glide, and Surflex rely on it. These programs use DUD-E to find active compounds among inactive ones. The table below shows how DUD-E supports these tools:
Virtual Screening Program | Database Used | Number of Targets | Average Ligands per Target | Decoys per Ligand |
|---|---|---|---|---|
Gold | DUD-E | 102 | 224 | 50 |
Glide | DUD-E | 102 | 224 | 50 |
Surflex | DUD-E | 102 | 224 | 50 |
FlexX | DUD-E | 102 | 224 | 50 |
Using datasets like LIT-PCBA avoids old dataset problems. It ensures your virtual screening results are fair and correct.
How LIT-PCBA Ensures Unbiased Screening
The LIT-PCBA dataset is special because it avoids bias. Older datasets often have repeated or overlapping data. LIT-PCBA fixes this by carefully choosing its content. This reduces mistakes and gives more realistic results.
LIT-PCBA focuses on single protein targets with X-ray structures. This focus helps make accurate predictions for experiments. It also balances active and inactive compounds. This balance helps train machine learning models without errors.
With LIT-PCBA, you can test your tools and improve them. It is a great resource for researchers working on drug discovery.
Applications of LIT-PCBA in Drug Discovery
The LIT-PCBA dataset is very useful for finding new medicines. Its high-quality data works well for many drug research tasks. For example, it helps test virtual screening methods for proteins like ESR1, KAT2A, MAPK1, and ALDH1. The table below explains these uses:
Dataset | Description |
|---|---|
LIT-PCBA(ESR1) | Made for virtual screening and machine learning. It uses 149 PubChem bioassays and removes false positives. It keeps similar properties for active and inactive compounds. |
LIT-PCBA(KAT2A) | Like ESR1, it is used for testing virtual screening. It ensures fair data to improve drug discovery accuracy. |
LIT-PCBA(MAPK1) | Provides a reliable dataset for virtual screening. It focuses on single protein targets with X-ray structures. |
LIT-PCBA(ALDH1) | Fixes problems in older datasets. It helps test virtual screening methods with better and more realistic data. |
By using LIT-PCBA, you can discover new ways to create medicines. Whether testing tools or making new ones, this dataset gives fair and useful data. It has already helped drug research and continues to inspire new ideas.
How LIT-PCBA Helps Machine Learning
Using Ligand Similarity to Train Models
The LIT-PCBA dataset helps improve machine learning models. It uses ligand similarity to tell active compounds from decoys. This is important for making accurate drug predictions. The dataset’s relaxed rules for finding active compounds make it a tough test. This ensures models are tested in realistic ways, leading to better training.
By focusing on ligand similarity, the dataset helps make models more precise. For example, studying active and inactive compounds in LIT-PCBA gives a strong base for training. This method helps machine learning tools find useful patterns, making them better for drug research.
Learning About Model Performance and Generalization
The LIT-PCBA dataset shows how well models work and adapt to tasks. Metrics like AUROC (Area Under the Curve) and EF1% (Enrichment Factor at 1%) measure model strengths and weaknesses. For example:
The EnOpt model had a high AUROC of 0.745, beating older methods like ensemble-average (0.588) and ensemble-best (0.599).
The ConPLex model, however, had a low AUROC of 50.9%, close to random guessing.
These results show how the dataset challenges models and reveals their abilities. Using LIT-PCBA helps you see where models do well and where they need work, improving real-world use.
Solving Problems in Machine Learning for Drug Research
Machine learning in drug research has problems like bias, bad data, and weak models. The LIT-PCBA dataset fixes these with good, unbiased data. Its design lowers overfitting risks and tests models in realistic ways.
Different models prove the dataset’s value in improving machine learning. For example:
Model Type | Description | Performance Metrics Used |
|---|---|---|
Meta-models | Mixes scoring functions with sequence-based deep learning. | Top5 recall, Top10 recall, Precision |
Ensemble models | Better than single models, with less bias and more stability. | Competes well with deep learning models |
Hybrid approaches | Combines physical and data-driven models for better results. | Tested on CASF-2016, PDBbind v2020, LIT-PCBA |
These models, tested on LIT-PCBA, show it can separate active and inactive compounds well. By solving common issues, the dataset helps create better machine learning tools for drug discovery.
Broader Impact and Future Applications of LIT-PCBA
Contributions to Open Science and Collaboration
The LIT-PCBA dataset helps open science grow. It gives researchers a trusted tool for virtual screening. Its activity labels come from reliable bioassays, making it accurate. This helps machine learning models work better and stay fair.
Older datasets had problems like bias and repeated data. LIT-PCBA fixes these issues, improving research results. For example, the SPRINT model tested with LIT-PCBA did better than others. It scored higher in AUROC, BEDROC, and enrichment factors.
Metric | SPRINT Model | Competitor Methods |
|---|---|---|
AUROC | Much higher | Lower |
BEDROC (alpha=0.85) | Much higher | Lower |
Enrichment Factor | Better at all levels | Lower |
Using LIT-PCBA connects you with scientists improving drug discovery.
Enhancing AI-Driven Drug Discovery
LIT-PCBA has changed how AI helps find new drugs. It supports advanced tools to find good compounds faster. For example, computer-aided drug design (CADD) uses LIT-PCBA to study molecules. It focuses on targets like PPAR gamma to find the best compounds.
Methodology | Description |
|---|---|
Virtual screening | Finds molecules that bind well to targets. |
Structural analogs screening | Tests compounds like GW9662 for medical use. |
Ligand-based screening | Uses filters and toxicity checks to improve results. |
Molecular docking | Tests binding strength using tools like iGemdock. |
These methods, powered by LIT-PCBA, have found better compounds faster.
Future Directions for Dataset Development
LIT-PCBA will grow to include more proteins and bioassays. This will make it even more useful for drug research. Better data curation will reduce errors and bias, keeping results strong.
In the future, LIT-PCBA may work with AI tools for better predictions. This will help study complex molecular interactions more precisely. These updates will keep LIT-PCBA leading in drug discovery and computational biology.
The LIT-PCBA dataset is important for virtual screening and machine learning. It helps study ligand similarity and checks performance on many targets. These features show how compounds interact and improve screening methods. With good data, it makes research more accurate and useful.
This dataset has changed drug discovery by fixing bias and bad data issues. Its smart design helps make big steps in computational biology. It also opens doors for future AI tools. Using LIT-PCBA, you can find new ways to create medicines faster and save lives.
FAQ
What makes the LIT-PCBA dataset special?
The LIT-PCBA dataset is unique because it avoids bias. It uses high-quality data that feels like real experiments. It focuses on single protein targets with X-ray structures. This makes it a trusted tool for virtual screening and finding new medicines.
How does LIT-PCBA help machine learning models?
LIT-PCBA trains models with balanced data and ligand similarity. This reduces mistakes and prevents overfitting. It also tests models with real-world challenges. This helps create better tools for drug research.
Can beginners work with the LIT-PCBA dataset?
Yes, beginners can use LIT-PCBA easily. Its clear structure and simple metrics make it beginner-friendly. Start by exploring its bioassays and testing basic virtual screening models. This will help you learn about drug discovery.
Why is unbiased data important in virtual screening?
Unbiased data gives fair and accurate results. It stops models from learning wrong patterns caused by repeated data. Using unbiased datasets like LIT-PCBA helps you trust your results and improve drug research.
What are some uses of the LIT-PCBA dataset?
You can use LIT-PCBA to test virtual screening tools and train models. It helps find new drug candidates faster. Tasks like molecular docking, ligand screening, and structural analysis are supported, speeding up drug discovery.
See Also
Understanding PCBA Datasets for Effective Defect Detection
Exploring the Significance of PCBA in Electronics
Defining PCBA: An Overview of Its Importance